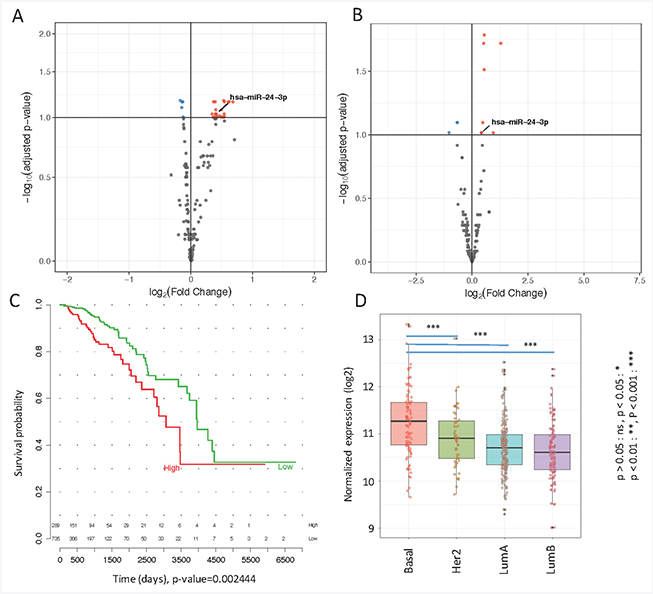
Researchers from NYU discuss their 2018 study published in Oncotarget, entitled, “Prognostic role of elevated mir-24-3p in breast cancer and its association with the metastatic process.”

Behind the Study is a series of transcribed videos from researchers elaborating on their recent oncology-focused studies published in Oncotarget. A new Behind the Study is released each Monday. Visit the Oncotarget YouTube channel for more insights from outstanding authors.
—
Francisco Esteva
Hi, my name is Francisco Esteva, Professor of Medicine and Oncology at NYU Langone Health at the Perlmutter Cancer Center. And we would like to tell you a little bit about the recent publication in Oncotarget, about the prognostic role of mir-24-3p in breast cancer and its association with the metastatic process. The motivation to do this project was to understand and identify biomarkers that will be associated with prognosis in breast cancer patients.
So we started with a prospective collection of plasma samples from patients with stage one to stage three breast cancer, and we followed those patients for a number of years. And we identified a group of patients who subsequently developed metastatic breast cancer. And then we found another group of patients who had not developed breast cancer, as a sort of a case control approach. And then we were interested in noncoding RNAs, particularly microRNAs. So we decided to use NanoString technology to look at a wide number of microRNAs and found one that we felt was very interesting. And we’ll discuss in more detail how we did it in the laboratory.
I’m a believer in the importance of translational research and how to apply information from genomics and gene sequencing studies to the clinic. And we can review a little bit what will be the clinical implications of this work. So I like Adriana to introduce herself and discuss some of the technology we used at NYU.
Adriana Heguy
Hi, my name is Adriana Heguy. I’m the Director of the Genome Technology Center, and recently have also become the Assistant Dean for all the Advanced Research Technologies here at NYU Langone. And the scope of our work is to provide really advanced technologies for physician scientists or basic scientists to be able to tap into this resources for translational projects, basic science projects. And this one was a perfect example of integration of clinical data, genomics data, bioinformatics data.
It was particularly challenging because these are plasma samples and it’s very easy, or a lot easier, to do research on tissues where the nucleic acids or the biomarkers are abundant. But for plasma samples, it is very challenging. And we took advantage of this technology called NanoString encounter technology that measures the microRNAs or the mRNAs directly without any PCR amplification. And in this case, you can use very small volumes of plasma to do the analysis, and because there’s no PCR amplification, there’s also less bias.
So it was an excellent technology for us to apply, but of course, at first we had many samples and we weren’t exactly sure how to extract the RNA. We had to work the perfect conditions. We also added spike-in exogenous microRNAs for us to be able to measure precisely the concentration of the real microRNAs and make sure we weren’t introducing any biases in the collection or the extraction of the RNA or the processing of the samples. And NanoString has put together a panel of 800 of these microRNAs. And so we can look at all 800 at once with a very little amount of total RNA. So this was really an ideal technology to look at this type of samples.
Alireza Khodadadi
Hi, my name is Alireza Khodadadi. I’m a Senior Bioinformatics Analyst at NYU Applied Bioinformatics Laboratories and Genome Technology Center at the Division of Advanced State Research Technologies. And in this paper we really wanted to use detecting these microRNAs, not only to just prognosis, but we’d wanted to put emphasis on … but not only for diagnosis, but our emphasis was on prognosis to find out which of the patients might in the future develop metastasis. And we also performed tests to identify the targets of this microRNA, to see the genes that are being targeted by this microRNA, what are the impacts of those and what roles they play in cancer.
Francisco Esteva
So one of the most interesting aspects of this research is to integrate real patient samples that we can collect prospectively in our clinics with information already available from The Cancer Genome Atlas (TCGA), which Reza analyzed extensively. More than a thousand patients with RNA sequencing data are publicly available and looking at the expression of these microRNAs in tissue samples from TCGA combined with our innovative approach at the looking at plasma microRNA using NanoString technology. And in this case combined, these are very powerful techniques to identify new biomarkers, which may have important implications in the future, since what we are trying to predict is which patients with early stage breast cancer will develop metastasis at some point in their lives, which could be of course, major event.
So I’d like Reza to comment a little bit more on the TCGA analysis, how he was able to mine the TCGA to find genes, in this case, microRNAs associated with metastasis and how then we went back to our patient samples to come up with this biomarker we identified, miR-24-3p in breast cancer.
Alireza Khodadadi
So the idea was to use TCGA primary tumor samples because cancer cells are expressing some of the genes abnormally high, and the product of these genes, whether proteins or RNAs, they leak into the blood. So we were thinking that the microRNA that we found in blood, that could be used for prognosis, is originating from a primary tissue. So we use the samples from TCGA and then ran statistical analyses to match the data and see if the microRNA that we found in the blood was originating from the primary tissues.
Francisco Esteva
And also finding pathways that may be associated with the mechanism of resistance. So in summary, this collaboration between clinicians, basic research, very sophisticated gene technology laboratory and bioinformatics, I think, is a good example of translational research and how we would like to move in the future of NYU Langone Health. Thank you.
Alireza Khodadadi
Thank you.
Adriana Heguy
Thank you.
Click here to read the full study published in Oncotarget.
—
Oncotarget is a unique platform designed to house scientific studies in a journal format that is available for anyone to read—without a paywall making access more difficult. This means information that has the potential to benefit our societies from the inside out can be shared with friends, neighbors, colleagues and other researchers, far and wide.
For media inquiries, please contact media@impactjournals.com.